
The Multi-locus ANTIgenic Simulator (MANTIS) software package was designed for the R statistical environment by the Evolutionary Ecology of Infectious Disease (EEID) research group of the Department of Zoology at the University of Oxford. MANTIS is a tool to simulate and analyse epidemiological time-series, generated under the biological assumptions of the strain theory of host-pathogen systems by Gupta et al 1998.
Abstract from publication in BMC Bioinformatics
In host-pathogen systems the development of immunity by the host places pressure on pathogens,
by setting up competition between genetic variants due to the establishment of cross-protective responses. These
pressures can lead to pathogen-specific, ubiquitous dynamic behaviours. Understanding the evolutionary forces that
shape these patterns is one of the key goals of computationally simulated epidemiological models. Despite the
contribution of such research methods in recent years to our current understanding of pathogen evolution, the
availability of free software tools for the general public remains scarce.
We developed the Multi-locus ANTIgenic Simulator
(MANTIS) software package for the R statistical
environment. MANTIS can simulate and analyse epidemiological time-series generated under the biological
assumptions of the strain theory of host-pathogen systems by Gupta et al.
MANTIS wraps a C/C++ ordinary-differential equations system and Runge-Kutta solver into a set of
user-friendly R functions. These include routines to numerically simulate the system and others to analyse, visualize
and export results. For this, the package offers its own set of time-series plotting and exportation functions. MANTIS's
main goal is to serve as a free, ready-to-use academic software tool. Its open source nature further provides an
opportunity for users with advanced programming skills to expand its capabilities. Here, we describe the background
theory, implementation, basic functionality and usage of this package.
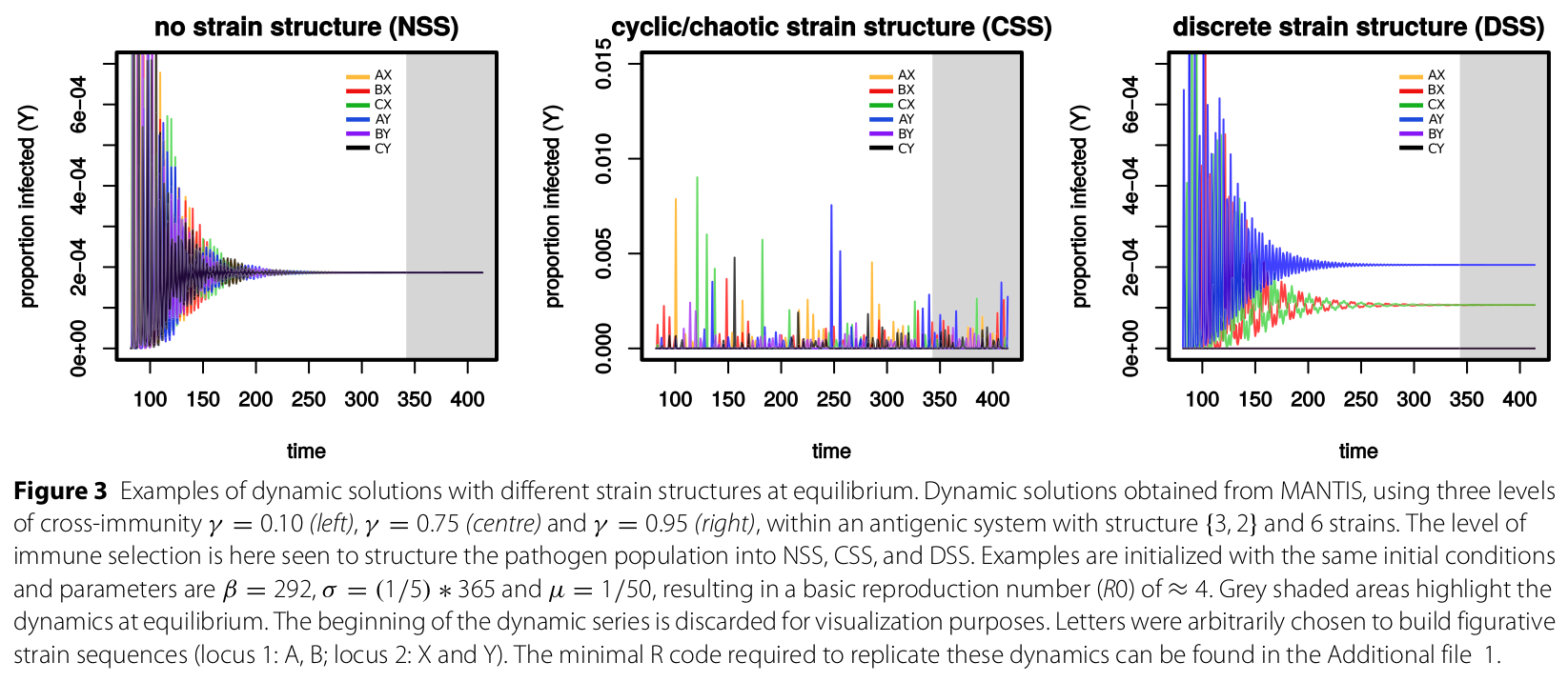
Summary of this figure: This figure presents examples of the three universal dynamic behaviours of multi-strain pathogens as predicted by strain theory. On the left, no strain structure (NSS), in which low or no cross-immunity between circulating strains allows all to be equally sucessful at the population level. In the middle, intermediate cross-immunity between strains keeps them in a transient dynamic behaviour termed cyclical or chaotic strain structure (CSS). On the right, strong or complete cross-immunity forces the strains to self organize into discrete subsets of antigenic types - discrete strain structure - in order to relax competition and able to circulate. Each subplot was simulated using the MANTIS R-package. More details in the academic publication
Availability and requirements
MANTIS has a dedicated manual that follows the well
established format for R packages. It presents a general
description of the package’s functionality, as well as examples,
a short introduction and key literature references for
strain theory. As new versions of the
package are expected in the near future, the reader should
refer to this manual for updated references and mathematical formulations of MANTIS's inbuilt methods.
The package can be installed using a platform-independent, source-based file (note: compiling source packages may require the pre-installation of Rtools on Windows and Xcode on Mac machines).
Both the manual and source file can be found at SourceForge.

under the GNU GPL 3.0 license. Detailed technical features of the
package can be found in the academic publication.