
In my research group, we have recently shown that a mosquito-borne viral suitability measure we called the index P can be derived from the R0’s expression of a climate-driven transmission dynamic model we have applied to Zika [a] and dengue [b]. In theory, the index P can be used as proxy for timing and scale of mosquito-borne viral transmission potential, without the need to estimate this from epidemiological data (e.g. number of cases in time). The index P's biological interpretation is the reproductive (transmission) potential of an adult female mosquito. The first demonstration of the index P was in the context of dengue in Myanmar [c].
The Mosquito-borne Viral Suitability index Estimator (MVSE) is an R-package that implements a vast series of functions which allow to simulate the index P for a particular virus, host, mosquito and region of interest. MVSE requires climatic variables as input, as well as some prior knowledge on key entomological and epidemiological parameters.
A research article demonstrating the potential of MVSE at multiple spatio-temporal scales in Brazil and Honduras is under review in Methods in Ecology and Evolution. Research article can be found here.
References: [a] Epidemiological and ecological determinants of Zika virus transmission in an urban setting. eLIFE 2017. Read it here. [b] Genomic and epidemiological characterisation of a dengue virus outbreak among blood donors in Brazil. Nature Sci. Rep. 2017. Read it here. [c] Measuring Mosquito-borne Viral Suitability in Myanmar and Implications for Local Zika Virus Transmission. PLoS Currents 2018. Read it here.
Abstract from publication
Viruses such as dengue, Zika, yellow fever and chikungunya depend on mosquitoes for transmission.
Their epidemics typically present periodic patterns, linked to the underlying mosquito population
dynamics, which are known to be driven by natural climate fluctuations. Understanding how climate
dictates the timing and potential of viral transmission is essential for preparedness of public
health systems and design of control strategies. While various alternative approaches have been
proposed to estimate local transmission potential of such viruses, few open-source, ready to use
and freely available software tools exist.
We developed the Mosquito-borne Viral Suitability Estimator (MVSE) software package for the
R programming environment. MVSE estimates the index P,
a novel suitability index based on a climate-driven mathematical expression for the basic
reproductive number of mosquito-borne viruses. By accounting for local humidity and temperature,
as well as viral, vector and human priors, the index P can
be estimated for specific host and viral species in different regions of the globe.
We describe the background theory, empirical support and biological interpretation
of the index P. Using real world examples
spanning multiple epidemiological contexts, we further demonstrate MVSE's basic
functionality, research and educational potentials
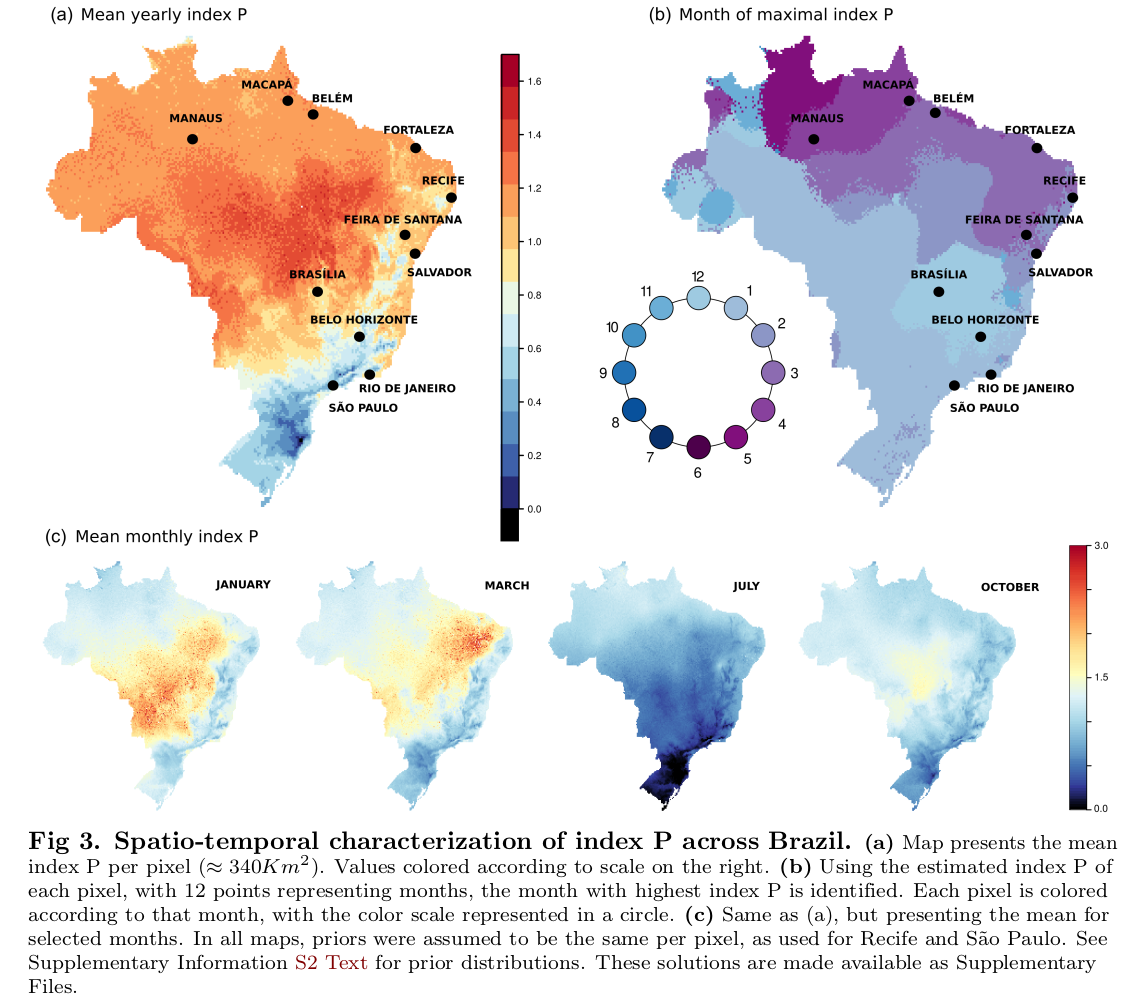
Summary of this figure: Subplot (a) of this figure presents the mean yearly index P (transmission potential) of dengue accross Brazil when using World Clim v2 data (climatic data with resolution of ~340 Km2 per pixel). Subplot (b) shows the month in which the index P peaks when using the same World Clim v2 data. Subplot (c) presents snapshots in time of the index P estimations using the same World Clim v2 data.
Availability and requirements
UPDATE: The current version of MVSE is 1.01r. The link below to SourceForge takes to this final version. To access previous versions please explore the SourceForge page.
The R-package MVSE is available under a GNU GPL 3.0 license at SourceForge.

There, you can find both a source-based package file and PDF reference manual can be found.
Newer versions and related materials will be deposited in such repository and
we refer the reader to it for further information and future changes.
MVSE is platform-independent requiring R (>= 3.4) and R-packages pbapply
(for user-console feedback), scales (for plotting), genlasso (for time series smoothing).
The package can be installed using a platform-independent, source-based file
(note: compiling source packages may require the pre-installation of Rtools
on Windows and Xcode on Mac machines).
Detailed technical features of the package can
be found in the academic publication,
including details on the climate-driven functions for ento-epidemiological
parameters, and priors used in the main text.